Assistant Professor Yohei Ishibashi and his research team have enable flexible production of polyunsaturated fatty acids in marine eukaryotic microorganisms through genome editing
Microbial production of diverse valuable fatty acids achieved without the use of transgenes
Points
- Docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA), classified as polyunsaturated fatty acids (PUFAs)*¹, are beneficial lipids that contribute to human health. However, supply sources that rely on natural resources face sustainability challenges, and there is a growing need to establish stable, safe, and secure alternative sources
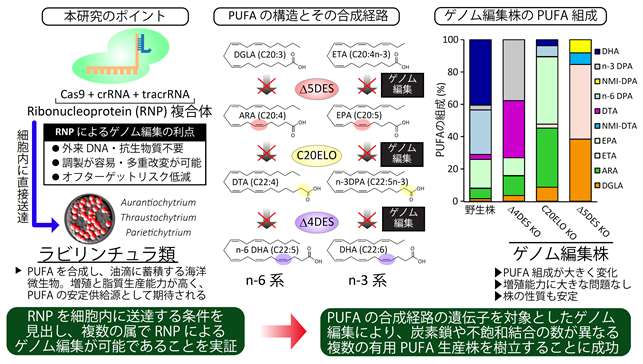
- Focusing on marine eukaryotic microorganism, thraustochytrids*², we developed a system for producing various valuable PUFAs using a genome editing method that does not require foreign DNA such as antibiotic resistance genes
- This achievement is expected to contribute to the improvement of thraustochytrid strains and serve as a fundamental technology for the industrial production of valuable PUFAs and the production of rare PUFAs*³
Abstract
Highly (poly) unsaturated fatty acids (PUFAs), represented by docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA), are beneficial lipids that contribute to human health. PUFAs are thought to exhibit different functions depending on the number of carbon atoms and the number and position of unsaturated bonds in their molecules. Thraustochytrids are marine eukaryotic microorganisms that biosynthesize and accumulate PUFAs at high levels. We have reported that one genus, Parietichytrium, possesses a complete pathway to synthesize DHA (22 carbons, 6 double bonds) as the final product through reactions involving multiple fatty acid elongases and desaturases, starting from palmitic acid (a saturated fatty acid with 16 carbons) (press release in 2021).
In this study, a research group consisting of Assistant Professor Yohei Ishibashi, Professor Nozomu Okino, and Professor Emeritus Makoto Ito of the Faculty of Agriculture, Kyushu University, along with Mr. Ryuji Tanimura and Mr. Yusuke Ataka (graduate students at the time of research) from the Graduate School of Bioresource and Environmental Sciences, and graduate student Akito Kumagai, in collaboration with Professor Daiske Honda of the Faculty of Science and Engineering, Konan University, demonstrated for the first time that genome editing technology*⁴ without the use of foreign DNA can be applied to thraustochytrids. In this study, without using any antibiotic resistance genes employed in previous research, we constructed a system capable of "tailored production"*⁵ of various PUFAs with different carbon numbers and degrees of unsaturation by modifying the PUFA biosynthetic pathway of the genus Parietichytrium. Currently, the main sources of DHA and EPA are fish oils derived from marine fish, but due to declining fish catches, concerns about climate change, and growing global demand for PUFAs, there is an urgent need to establish more stable, safe, and secure alternative sources. This research is expected to contribute to the realization of sustainable industrial production of PUFAs without relying on natural resources. Furthermore, it enables the production of "rare PUFAs" that are scarcely present in natural resources such as fish oil. These rare PUFAs may possess beneficial functions that have previously been overlooked. In the future, we aim to elucidate the functions and verify the usefulness of these rare PUFAs. In addition, the genome editing technology without foreign DNA used in this study was confirmed to be applicable not only to the genus Parietichytrium but also to other genera, such as Aurantiochytrium, which is already being utilized industrially. As a breeding technology that further expands the potential of thraustochytrids as a stable source of valuable lipids, this approach is expected to have wide-ranging applications.
This research was published online in Chemical Engineering Journal, an Elsevier journal, on November 28, 2025 (Japan time).
Message From Our Research Group
We reported in 2021 that the genus Parietichytrium possesses all the genes encoding fatty acid elongases and desaturases involved in PUFA biosynthesis. The present achievement realizes the concept we had envisioned at that time--metabolic pathway modification through genome editing. We expect that the developed method and the genome-edited strains obtained will contribute to the advancement of research on thraustochytrids and lead to future applications.
Glossary
*1 highly (poly) unsaturated fatty acids (PUFAs)
Fatty acids that contain two or more cis-configured double bonds separated by a single methylene group (figure 1). PUFAs with a carbon chain length of 20 or more are called long-chain PUFAs (LC-PUFAs), and this study mainly focuses on LC-PUFAs. PUFAs are classified as either n-3 or n-6 series based on the position of the unsaturated bond closest to the methyl end (figure 1). They are thought to exhibit different functions depending on the number of carbon atoms and the number and position of unsaturated bonds.
Docosahexaenoic acid (DHA, C22:6) and eicosapentaenoic acid (EPA, C20:5n-3) are well-known n-3 PUFAs used as supplements and pharmaceuticals.
*2 Thraustochytrids
Marine eukaryotic microorganisms that synthesize PUFAs and accumulate them at high levels in oil droplets. Phylogenetically, they are closely related to algae, which include diatoms and brown algae such as kelp. Industrial utilization has advanced for one genus, Aurantiochytrium (sometimes referred to by the genus name Schizochytrium). Thraustochytrids synthesize PUFAs through three distinct types of pathways:
① a type that directly synthesizes DHA from precursors using a polyketide-like enzyme complex called PUFA synthase; ② a type that employs fatty acid elongases (ELO) and desaturases (DES) stepwise to synthesize DHA via various intermediate fatty acids with different chain lengths and degrees of unsaturation (figure 1); ③ a type that uses both pathways. These three types were identified in our previous study (Ishibashi et al, Commun. Biol. 2021). In this study, we demonstrated that genome editing without foreign DNA is applicable to three different genera: Aurantiochytrium (type ①), Parietichytrium (type ②), and Thraustochytrium (type ③). This finding highlights the broad applicability of this approach across thraustochytrids.
*3 rare PUFAs
In this study, "rare PUFAs" are defined as PUFAs that are present only in trace amounts or almost absent in natural resources, unlike PUFAs such as DHA and EPA that are abundant in fish oil and other natural sources. Examples include ETA produced in strains lacking the Δ5DES gene, and DTA or n-3DPA obtained from strains lacking the Δ4DES gene (figure 1). Furthermore, this paper reveals that in Δ5DES-deficient strains, sites that would normally undergo desaturation remain unchanged, allowing subsequent reactions to proceed and resulting in the synthesis of non-methylene-interrupted PUFAs (NMI-PUFAs) (figure 1). These rare PUFAs and NMI-PUFAs may possess functions and benefits distinct from those of DHA and EPA. To elucidate these functions, sufficient quantities of rare PUFAs are required for validation using model organisms; however, their low abundance in natural resources makes procurement difficult. By using the genome-edited strains reported in this study, these rare PUFAs can be produced stably and in the necessary amounts, contributing to the advancement of research aimed at verifying their functions.
*4 genome editing technology
A general term for techniques that modify the DNA sequence of an organism at targeted locations. A representative method is "CRISPR-Cas9," which was also used in this study. By precisely and efficiently cutting and repairing specific genes, genome editing is widely applied to the improvement of crops, aquatic resources, and industrial microorganisms. In this study, we examined a genome editing approach without foreign DNA, using only ribonucleoproteins (RNPs). Specifically, we prepared RNPs composed of Cas9 protein and guide RNAs (crRNA + tracrRNA), delivered them directly into cells via electroporation, and selected strains in which the target gene was modified. This method avoids the insertion of foreign DNA, such as antibiotic resistance genes, into the genomic DNA of the target organism, thereby preventing genetic recombination. It also eliminates limitations associated with the number of selectable markers and the additional steps required to reuse markers in conventional genetic engineering. Furthermore, because RNPs are degraded inside cells, this approach is expected to reduce off-target effects caused by sustained Cas9 expression. In fact, the genome-edited strains established in this study showed no reduction in growth or PUFA production and accumulation capacity, suggesting that off-target effects problematic for PUFA-producing microorganisms did not occur.
*5 tailored PUFA production
Aurantiochytrium grow rapidly and exhibit high PUFA production and accumulation capacity, making them widely used in industry. However, the types of PUFAs they produce are limited to DHA and n-6 docosapentaenoic acid (n-6DPA), which are products of PUFA synthase. In contrast, Parietichytrium convert various fatty acids with different carbon chain lengths and degrees of unsaturation into DHA through a stepwise pathway. By knocking out specific enzyme genes in this pathway, it is possible to halt the process at upstream intermediates (figure 1). In this study, genome editing targeted the Δ4 desaturase (Δ4DES) gene to create strains (Δ4DES knockout strains) that synthesize n-3DPA instead of DHA and docosatetraenoic acid (DTA) instead of n-6DPA as their major PUFAs (figure 1). Similarly, knocking out the C20 elongase (C20ELO) gene enabled the production of EPA and arachidonic acid (ARA), while knocking out the Δ5 desaturase (Δ5DES) gene resulted in strains producing eicosatetraenoic acid (ETA) and dihomo-γ-linolenic acid (DGLA) (figure 1). Using these genome-edited Parietichytrium strains, it became possible to produce valuable PUFAs other than DHA and n-6DPA at high yields through microbial fermentation.
Publication Information
Journal: Chemical Engineering Journal
Title: Transgene-Free Protein-Based Genome Editing in Thraustochytrids Enables Customizable Modulation of Long-Chain Polyunsaturated Fatty Acid Profiles
Authors: Yohei Ishibashi*, Ryuji Tanimura, Yusuke Ataka, Akito Kumagai, Daiske Honda, Makoto Ito, Nozomu Okino
DOI:10.1016/j.cej.2025.171156
- For more details on this research, click here.
For Research-related inquiries











 Contact
Contact
 Access Map
Access Map

