Assistant Professor Takamasa Teramoto, Professor Yoshimitsu Kakuta, and Their Research Group Uncover the Mechanism Behind Dual-End Cleavage in Transfer RNAs
Dual-Function Strategy Uncovered in a Compact Enzyme: A Star-Shaped RNase P Complex Precisely Processes Both Ends of Precursor tRNA
A research team led by Assistant Professor Takamasa Teramoto and Professor Yoshimitsu Kakuta at Kyushu University's Laboratory of Biophysical Chemistry (Division of Applied Biosciences) has uncovered a novel molecular mechanism by which a compact ribonuclease enzyme achieves precise dual-end cleavage of precursor tRNA. Their findings reveal a unique evolutionary strategy wherein a small enzyme complex acquires additional functionality through oligomerization.
Key Findings
- Transfer RNA (tRNA) must undergo precise processing at both its 5′ and 3′ ends to become functional. While the mechanism of 5′-end cleavage has been well studied, how certain bacteria achieve accurate trimming at both ends remained unclear.
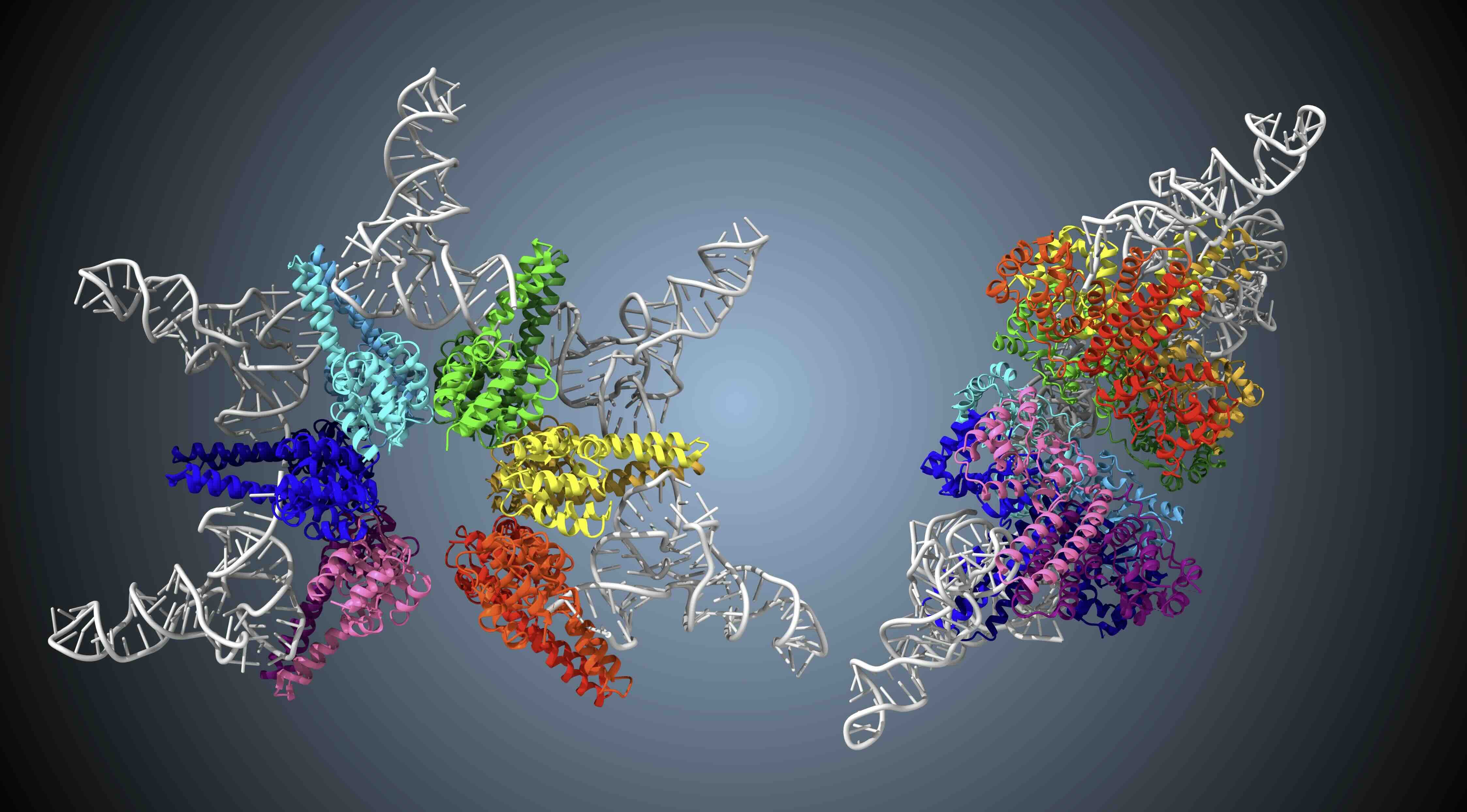
- The team discovered that a minimalist form of ribonuclease P (RNase P)--previously thought to only cleave the 5′ end--assembles into a 12-subunit, star-shaped complex with six radial protrusions. This architecture cradles the precursor tRNA between protrusions, enabling the enzyme to cleave not only the 5′ end but also the 3′ end.
- This is the first demonstration of such a "dual-function" mechanism for RNase P, expanding its known biological role. The findings illustrate a previously unrecognized evolutionary strategy whereby small enzymes gain new functions--like 3′-end processing--through structural assembly, while preserving their essential role in 5′-end cleavage.
- This discovery sheds light on the molecular evolution of enzymes, showing how compact proteins can expand their functional repertoire through multimerization, a strategy that could have broader implications for understanding RNA processing and the design of biomolecular tools.
Summary
Using cryo-electron microscopy (cryo-EM), the team analyzed the structure of HARP and revealed that this enzyme assembles into a star-shaped dodecamer composed of 12 subunits. The structure specifically accommodates precursor tRNA and enables precise cleavage at the 5′ end. Furthermore, through structural and biochemical analyses, the team demonstrated for the first time that this compact enzyme also cleaves the 3′ end of precursor tRNA--uncovering a novel "dual-function" mechanism. This discovery not only provides new insight into the molecular evolution strategies of small enzymes but also holds promise for the development of compact RNA-cleaving molecular tools for RNA therapeutics, by engineering target RNAs into tRNA-like structures. Future studies are expected to elucidate the principles behind the assembly of this star-shaped complex and the regulation of its cleavage precision.
These findings were published in Nature Communications on [July 1st, 2025].
Figure for Reference
Publication Information
Journal: Nature Communications
Title: Structural basis of transfer RNA processing by bacterial minimal RNase P
Authors: Takamasa Teramoto†,, Takeshi Koyasu†, Takashi Yokogawa, Naruhiko Adachi, Kouta Mayanagi, Takahiro Nakamura, Toshiya Senda, Yoshimitsu Kakuta
†These authors contributed equally to this work.
*Corresponding authors
DOI: https://doi.org/10.1038/s41467-025-60002-1
Contact Information
Takamasa Teramoto, Assistant Professor
Yoshimitsu Kakuta, Assistant Professor
Laboratory of Biophysical Chemistry











 Contact
Contact
 Access Map
Access Map

